Events
Email Subscription
Subscribe to Filter
Importing into Google Calendar
In the left-hand column of the Google Calendar main view, click the arrow to the right of "Other calendars" and click "Add by URL". In the form that appears, paste in the URL from the box above, and click the button to confirm.
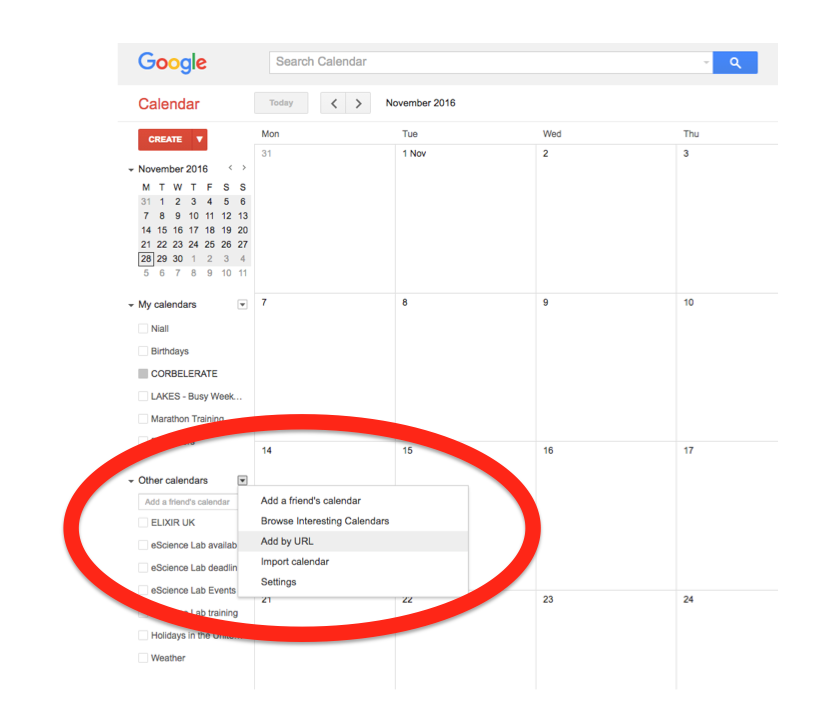
Please note, it may take a while for newly created events in TeSS to synchronise with your Google Calendar.
-
Face-to-faceWorkshops and coursesTraining Course on Best practices for RNA-Seq data analysis
27 - 29 September 2017
Fisciano, Italy
Face-to-face
Gene expression RNA-Seq Bioinformatics Sequencing RNA splicing Transcriptomics NGS Data analysis High Throughput Sequencing Analysis Gene Expression -
Face-to-faceGenome sequencing technologies, assembly, variant calling and statistical genomics
22 October - 2 November 2018
Oslo, Norway
Face-to-face
Genomics Bioinformatics Statistics and probability Sequencing genomics bioinformatics Genome sequence analysis -
Face-to-faceWorkshops and coursesCABANA workshop: Introduction to next-generation sequencing
18 - 20 March 2019
Mexico
Face-to-face
Sequencing -
Face-to-faceMeetings and conferencesRevolutionizing Next-Generation Sequencing (3rd edition)
25 - 26 March 2019
Antwerpen, Belgium
Face-to-face
Mapping Population genomics Population genetics Sequencing Transcriptomics Metagenomic sequencing Metagenomics Epigenetics Epigenomics Genotyping experiment Genotype and phenotype Genomics Genome Structure Genome Mapping Spatial transcriptomics Population Scale clinical sequencing Single Cell Sequencing -
OnlineWorkshops and coursesEuropean Nucleotide Archive: An introduction
3 April 2019 @ 09:00 - 17:00
Online
nucleotide Sequencing European Nucleotide Archive ENA Raw sequencing data Open-access data archive -
Face-to-faceWorkshops and coursesNext generation sequencing bioinformatics
23 - 26 September 2019
United Kingdom
Face-to-face
Sequencing Genomics DNA & RNA (dna-rna) -
Face-to-faceMeetings and conferencesThe Brain Mosaic: Cellular heterogeneity in the CNS (2nd edition)
10 - 11 October 2019
Leuven, Belgium
Face-to-face
Sequencing Transcriptomics Bioinformatics Single cell sequencing cell diversity Spatial transcriptomics transcriptomics Alzheimers Clinical Trials And StudiesAlzheimers DiseaseAmnesiaAnimal Models In DementiaAnxiety Autoimmune Disorders & Multiple SclerosisBehavioral IssuesBiopsychiatry Blood-Brain BarrierBrain DiagnosisBrain Disease & FailureBrain InjuryBrain TumourCentral Nervous SystemCerebrovascular DiseaseChild NeurologyClinical NeurophysiologyCNS TumorsCognitive DisordersDementiaDementia CareDemyelinationDiagnosis And TherapyEpilepsyGenetics Genetics And Epigenetics Geriatric NeurologyGut- Brain AxisHeadacheIntervention NeuroradiologyMeningitisMental HealthMental Health NursingMetastatisMovement DisordersMultiple SclerosisMultiple Sclerosis DiagnosisNervous SystemNeural EngineeringNeural EnhancementNeuro-Oncology NeuroanaatomyNeuroanatomyNeurocritical CareNeurodegenerationNeurodegenerative DisorderNeurodegenerative DisordersNeurogeneticsNeuroimagingNeuroimmunologyNeurological DisordersNeurological Disorders And StrokeNeurological InfectionsNeurological SurgeryNeurologistNeurologyNeuromedicinesNeuromuscular MedicineNeuronal AutoantibodiesNeuropathyNeuropeptidesNeuropharmacologyNeuroscienceNeurosurgeryNeurotoxicologyNeurotoxinsNeurotransmissionNeurotransmittersParkinsonParkinsons DiseasePediatric NeurologyPharmacology Psychiatry Psychology PsychoneuroimmunologyRecent Research And Case StudiesRehabilitation TherapyStem Cell Treatment For Neurological DisordersStrokesTraumatic Brain Injury CNS sequencing disease research bioinformatics • New developments in technology -
OnlineWorkshops and coursesSubmitting metagenomic data to ENA
21 November 2019 @ 09:00 - 17:00
Online
Metagenomic sequencing nucleotide Gene functional annotation Sequencing DNA & RNA (dna-rna) -
Face-to-faceWorkshops and coursesCABANA Workshop: NGS analysis applied to virome sequencing in agricultural systems
24 February - 4 March 2020
Costa Rica
Face-to-face
Sequencing Virology Experimental design and studies Genomics CABANA -
Face-to-faceWorkshops and coursesRNA-seq and single cell RNA-seq Course
9 - 13 March 2020
Torino, Italy
Face-to-face
RNA-Seq Sequencing
Note, this map only displays events that have geolocation information in
TeSS.
For the complete list of events in TeSS, click the grid tab.